DREAM Research Group
Laboratory for Applied Genome Technologies and Regenerative Medicine
New publication
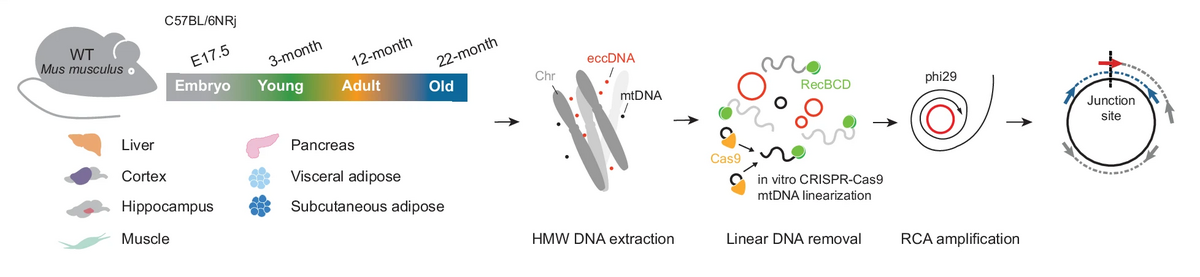
A new collaborative study from the DREAM group has recently been published in Nature Communications. This study illustrates the new roles of extrachromosomal circular DNA in shaping the architecture of genes and transcription activity in tissues.
A new collaborative study from the DREAM team is published in Nature Biotechnology. Link to full text.
In this new study, a cancer-specific long non-coding RNA (lncRNA, or calledTu-Stroma) was discovered in exosome from ovarian tomur cells. Tu-Stroma can specifically regulate the alternative splicing of the Flower gene, a gene which altertive splicing affect the cell fitness in tumor microenvironment. This discover has led to the development of a novel antibody-based therapy for tumor. 09 December 2024
Group leader Professor Yonglun Luo has received approximately DKK 5.7 million as part of a major EU-funded research project focused on animal welfare.
Read full news here. 4 December 2024 by Sebastian Alexander Skousgaard
Research raises hopes for new treatment of fusion-driven cancer
Researchers from Aarhus University have developed a new form of gene therapy that can stop cell division in a particularly aggressive type of cancer.
Link to full news 11 August 2023 by Line Rønn
 From left: Associate Professor Maja Ludvigsen, PhD student Signe Neldeborg, PhD student Johannes Frasez Sørensen, Professor Christian Kanstrup Holm, Associate Professor Magnus Stougaard and Professor Yonglun Luo. The researchers have studied the new form of gene therapy in a fusion-driven subtype of acute myeloid leukaemia (AML). The results provide hope for a completely new method of treatment specifically aimed at fusion-driven cancers for which there is currently no cure. Photo: Line Rønn
From left: Associate Professor Maja Ludvigsen, PhD student Signe Neldeborg, PhD student Johannes Frasez Sørensen, Professor Christian Kanstrup Holm, Associate Professor Magnus Stougaard and Professor Yonglun Luo. The researchers have studied the new form of gene therapy in a fusion-driven subtype of acute myeloid leukaemia (AML). The results provide hope for a completely new method of treatment specifically aimed at fusion-driven cancers for which there is currently no cure. Photo: Line Rønn
Two DANDRITE affiliated researchers receives Ascending Investigator-grant from the Lundbeck Foundation
Associate professor Marina Romero-Ramos and professor Yonglun Luo are among the 11 research talents to receive a grant of 5 million Danish kroner over a period of 4 years. The grant is earmarked for new research projects within the field of neuroscience and is meant to support researchers at the associate professor or professor level who want to cultivate new research areas within their scientific field.
Link to full news on 21 November 2022 by Rikke Skovgaard Lindhard
New professor aims to find cure for degenerative diseases in our cells
Newly appointed Professor Yonglun Luo from Aarhus University hopes to help patients with degenerative diseases such as Duchenne Muscular Dystrophy by unlocking a cure with his research.
Link to full news 1 November 2022 by Vibe Bregendahl Noordeloos
New study identifies functionally distinct blood endothelial cells in breast cancer
September 20, 2022
The dream team is part of a new study published in Nature Communications, which discovers a new type of blood endothelial cells with lipid processing functions in human breast (cancer) tissues. The new study constructed a EC taxonomy in breast (cancer) tissues, and identified clinically relevant EC phenotypes. This type of endothelial cells, called LIPEC, express genes involved in lipid processing and are more abundant in peri-tumoral breast tissues.
Link to the article:
Geldhof V, de Rooij LPMH, Sokol L, Amersfoort J, De Schepper M, Rohlenova K, Hoste G, Vanderstichele A, Delsupehe AM, Isnaldi E, Dai N, Taverna F, Khan S, Truong AK, Teuwen LA, Richard F, Treps L, Smeets A, Nevelsteen I, Weynand B, Vinckier S, Schoonjans L, Kalucka J, Desmedt C, Neven P, Mazzone M, Floris G, Punie K, Dewerchin M, Eelen G, Wildiers H, Li X, Luo Y, Carmeliet P. Single cell atlas identifies lipid-processing and immunomodulatory endothelial cells in healthy and malignant breast. Nat Commun. 2022 Sep 20;13(1):5511. doi: 10.1038/s41467-022-33052-y. PMID: 36127427.
The DREAM team releases its pig single cell transcriptome landscape results in Nature Communications
On June 24, 2022, the DREAM team, together with a large international collaboration group, releases its interim results from the Pig Atlas in Nature Communications. In the new study [1], the authors complete the single cell RNA sequencing of over 200,000 pig cells from 20 different tissues and organs. The new study provides interesting insights into blood endothelial cell heterogeneity and the evolutionally conserved gene expression network in brain residing microglia.
1. Wang, F., Ding, P., Liang, X., Ding, X., Brandt, C. B., Sjöstedt, E., Zhu, J., Bolund, S., Zhang, L., de Rooij, L., Luo, L., Wei, Y., Zhao, W., Lv, Z., Haskó, J., Li, R., Qin, Q., Jia, Y., Wu, W., Yuan, Y., … Luo, Y. (2022). Endothelial cell heterogeneity and microglia regulons revealed by a pig cell landscape at single-cell level. Nature communications, 13(1), 3620. https://doi.org/10.1038/s41467-022-31388-z
Direct URL to the Pig Single Cell Atlas: https://dreamapp.biomed.au.dk/pigatlas/
New study unravels the mechanisms affecting CRISPR/Cas9 activity and specificity
A new joint study contributed the DREAM team discovered that free binding energy changes and PAM context affect CRISPR/Cas9 activity and specificity. This finding leads to the development of better CRISPR design tools to select gRNAs with high efficiency and specificity. This study was published in Nature Communications on May 30, 2022.
Link to the article: https://www.nature.com/articles/s41467-022-30515-0
Link to news release by CRISPR Medicine News. https://crisprmedicinenews.com/news/new-study-explains-why-grna-efficiency-varies-at-on-and-off-target-sites/
Genome-wide annotation of protein-coding genes in pig
We are pleased to share our latest publication on the genome-wide annotation of protein-coding genes in pigs. This international collaboration project was innitated back in 2017 with an aim to better annotation the genome, gene expression, gene regultion and cell functions in pigs.
Visit the newly arrived pig atlas!
Publication: Karlsson, M., Sjöstedt, E., Oksvold, P. et al. Genome-wide annotation of protein-coding genes in pig. BMC Biol 20, 25 (2022). https://doi.org/10.1186/s12915-022-01229-y
NEW GRANT STRENGTHENS RESEARCH INTO CURING MUSCULAR DYSTROPHY
Associate Professor Yonglun Luo from Aarhus University receives DKK 2.7 million from the Novo Nordisk Foundation to study whether muscular dystrophy can be cured with the help of gene editing and stem cell technologies. The research is of particular benefit to patients with a rare muscular dystrophy disease.
2022.01.20 | LENE HALGAARD
https://newsroom.au.dk/en/news/show/artikel/ny-bevilling-styrker-forskning-i-muskelsvind/